Note
Click here to download the full example code
Basic NMA of a Protein-ENM¶
In this example script, a basic normal mode analysis (NMA) of a protein coarse-grained elastic network model (ENM) is conducted.
# Code source: Jan Krumbach
# License: BSD 3 clause
import numpy as np
import matplotlib.pyplot as plt
import biotite
import biotite.structure as struc
import biotite.structure.io.mmtf as mmtf
import biotite.database.rcsb as rcsb
import springcraft
# Fetch G:T/U Mismatch-specific DNA glycosylase from E. coli
PDB_ID = "1MUG"
mmtf_file = mmtf.MMTFFile.read(rcsb.fetch(PDB_ID, "mmtf"))
whole_structure = mmtf.get_structure(mmtf_file, model=1, include_bonds=True)
protein = whole_structure[struc.filter_amino_acids(whole_structure)]
ca = protein[(protein.atom_name == "CA") & (protein.element == "C")]
# Select forcefield; create ANM object using the eANM forcefield
ff = springcraft.TabulatedForceField.e_anm(ca)
eanm = springcraft.ANM(ca, ff)
## NMA
# Compute eigenvalues and eigenvectors.
# The first 6 eigenvals./eigenvecs corresponding to trivial modes are omitted.
# -> analyse modes 7-107
eigenval, eigenvec = eanm.eigen()
eigenval = eigenval[6:106]
# Compute fluctuations for all residues and frequencies for modes 7-107
msqf = eanm.mean_square_fluctuation()
freq = eanm.frequencies()[6:106]
## Plot
fig = plt.figure(figsize=(8.0, 8.0), constrained_layout=True)
grid = fig.add_gridspec(nrows=2, ncols=2)
ax00 = fig.add_subplot(grid[0, 0])
ax01 = fig.add_subplot(grid[0, 1])
ax1 = fig.add_subplot(grid[1, :])
biotite_c = biotite.colors["orange"]
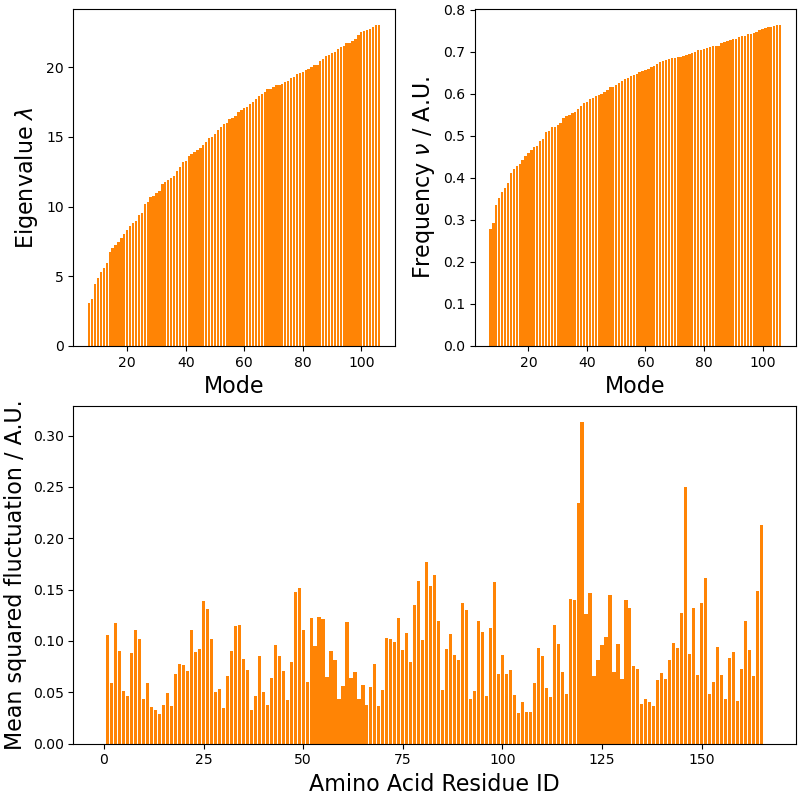
ax00.bar(x=np.arange(7, len(eigenval)+7), height=eigenval, color=biotite_c)
ax01.bar(x=np.arange(7, len(freq)+7), height=freq, color=biotite_c)
ax1.bar(x=np.arange(1, len(msqf)+1), height=msqf, color=biotite_c)
ax00.set_xlabel("Mode", size=16)
ax00.set_ylabel(r"Eigenvalue $\lambda$", size=16)
ax01.set_xlabel("Mode", size=16)
ax01.set_ylabel(r"Frequency $\nu$ / A.U.", size=16)
ax1.set_xlabel("Amino Acid Residue ID", size=16)
ax1.set_ylabel("Mean squared fluctuation / A.U.", size=16)
plt.show()
